Caracterización del genoma completo y análisis de microsatélites mediante un enfoque in silico en pavos domésticos (Meleagris gallopavo)
DOI:
https://doi.org/10.18633/biotecnia.v27.2455Palabras clave:
in sílico, pavos, motivos, microsatélites, Meleagris gallopavoResumen
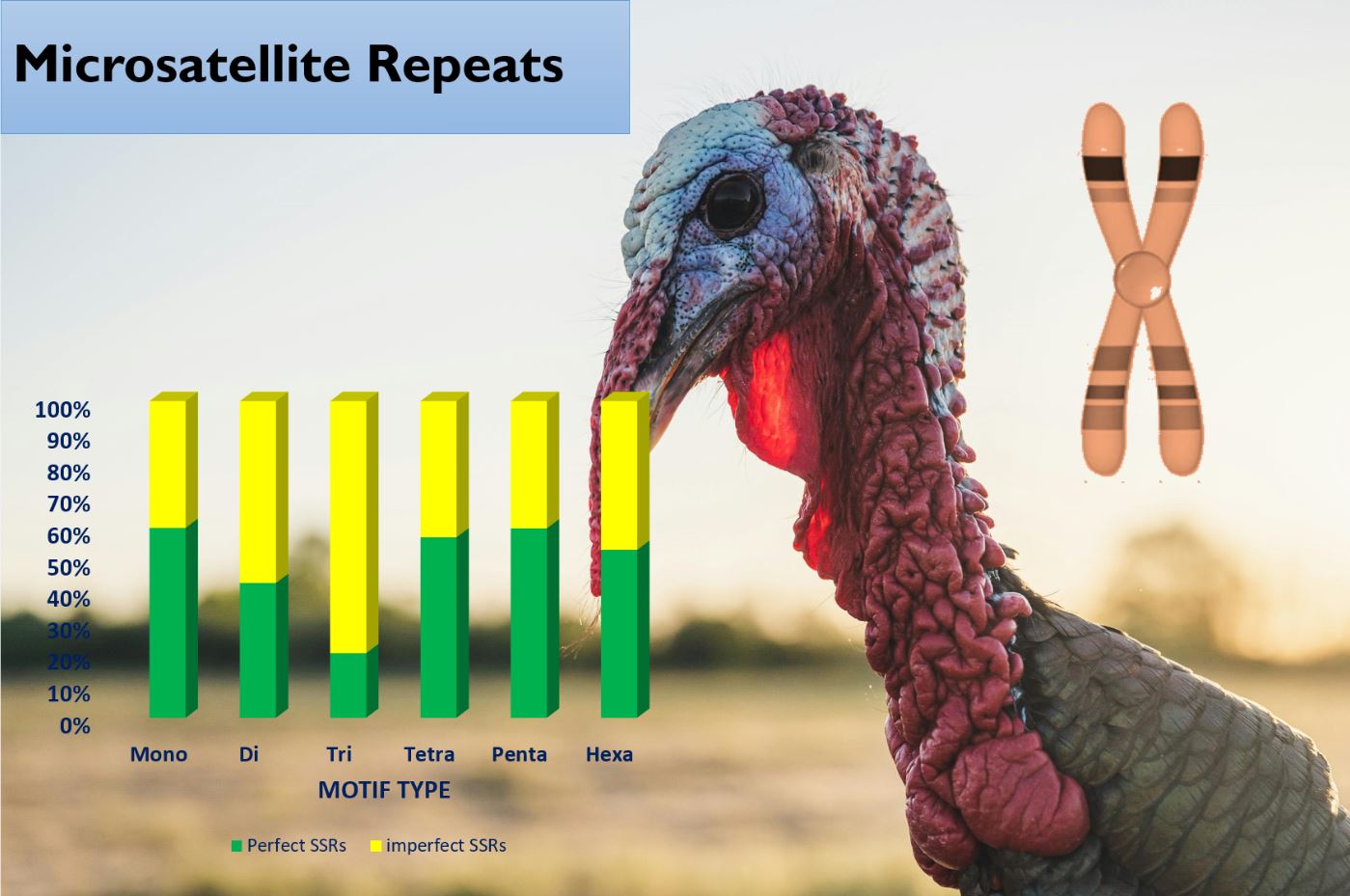
El pavo doméstico (Meleagris gallopavo) es el segundo mayor contribuyente a la producción de carne de aves de corral, después de los pollos. El estudio de la organización y distribución de microsatélites es muy importante en los estudios genómicos y evolutivos. La minería in silico de microsatélites aprovecha el poder de la biología computacional para agilizar, mejorar el descubrimiento de marcadores de microsatélites y reducir el costo de la detección de microsatélites. El presente estudio tuvo como objetivo evaluar la minería in silico de loci de microsatélites en el genoma del pavo doméstico. Se obtuvieron secuencias de referencia de varios cromosomas de NBCI y se analizaron utilizando el software Krait. El cromosoma 4 tuvo el mayor número de microsatélites perfectos, mientras que el cromosoma 18 tuvo el menor número. Sin embargo, el cromosoma 27 tuvo la mayor abundancia relativa, seguido por el cromosoma 13. El cromosoma 18 nuevamente tuvo la menor abundancia relativa. El cromosoma 4 tuvo la mayor cantidad de microsatélites imperfectos y el cromosoma 18 tuvo la menor cantidad. Se diseñaron un total de 121.248 cebadores de microsatélites. Estos loci y marcadores microsatélites desempeñarán un papel importante en el mapeo de ligamiento y mejorarán significativamente la investigación sobre la genética del pavo.
Descargas
Citas
Abdul-Muneer, P.M. 2014: Application of Microsatellite Markers in Conservation Genetics and Fisheries Management: Recent Advances in Population Structure Analysis and Conservation Strategies: Genetics Research International: 2014: 1–11: 10.1155/2014/691759.
Akemi, A., Pereira, J., Macedo, P., and Alessandra, K. 2012: Microsatellites as Tools for Genetic Diversity Analysis: Genetic Diversity in Microorganisms: 2: 64: 10.5772/35363.
Axelsson, E., Webster, M.T., Smith, N.G.C., Burt, D.W., and Ellegren, H. 2005: Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes: Genome Research: 15: 120–125: 10.1101/gr.3021305.
Barros, C.P., Derks, M.F.L., Mohr, J., Wood, B.J., Crooijmans, R.P.M.A., Megens, H.J., Bink, M.C.A.M., and Groenen, M.A.M. 2023: A new haplotype-resolved turkey genome to enable turkey genetics and genomics research: GigaScience: 12: giad051: 10.1093/gigascience/giad051.
Burt, D.W., Morrice, D.R., Sewalem, A., Smith, J., Paton, I.R., Smith, E.J., Bentley, J., and Hocking, P.M. 2003: Preliminary linkage map of the Turkey (Meleagris gallopavo) based on microsatellite markers: Animal Genetics: 34: 399–409: 10.1046/j.1365-2052.2003.01033.x.
Canales Vergara, A.M., Landi, V., Delgado Bermejo, J.V., Martínez, A.M., Cervantes Acosta, P., Pons Barros, A., Bigi, D., Sponenberg, P., Helal, M., Banabazi, M.H., and Camacho Vallejo, M.E. 2020: Design and development of a multiplex microsatellite panel for the genetic characterisation and diversity assessment of domestic turkey (Meleagris gallopavo gallopavo): Italian Journal of Animal Science: 19: 392–398: 10.1080/1828051X.2020.1745695.
Clauss, M., Frei, S., Hatt, J.M., and Kreuzer, M. 2020: Methane emissions of geese (Anser anser) and turkeys (Meleagris gallopavo) fed pelleted lucerne: Comparative Biochemistry and Physiology -Part A : Molecular and Integrative Physiology: 242: 110651: 10.1016/j.cbpa.2020.110651.
Dalloul, R.A., Long, J.A., Zimin, A. V., Aslam, L., Beal, K., Blomberg, L.A., Bouffard, P., Burt, D.W., Crasta, O., Crooijmans, R.P.M.A., Cooper, K., Coulombe, R.A., De, S., Delany, M.E., Dodgson, J.B., Dong, J.J., Evans, C., Frederickson, K.M., Flicek, P., Florea, L., Folkerts, O., Groenen, M.A.M., Harkins, T.T., Herrero, J., Hoffmann, S., Megens, H.J., Jiang, A., de Jong, P., Kaiser, P., Kim, H., Kim, K.W., Kim, S., Langenberger, D., Lee, M.K., Lee, T., Mane, S., Marcais, G., Marz, M., McElroy, A.P., Modise, T., Nefedov, M., Notredame, C., Paton, I.R., Payne, W.S., Pertea, G., Prickett, D., Puiu, D., Qioa, D., Raineri, E., Ruffier, M., Salzberg, S.L., Schatz, M.C., Scheuring, C., Schmidt, C.J., Schroeder, S., Searle, S.M.J., Smith, E.J., Smith, J., Sonstegard, T.S., Stadler, P.F., Tafer, H., Tu, Z., van Tassell, C.P., Vilella, A.J., Williams, K.P., Yorke, J.A., Zhang, L., Zhang, H. Bin, Zhang, X., Zhang, Y., and Reed, K.M. 2010: Multi-platform next-generation sequencing of the domestic Turkey (Meleagris gallopavo): Genome assembly and analysis: PLoS Biology: 8: e1000475: 10.1371/journal.pbio.1000475.
Ding, S., Wang, S., He, K., Jiang, M., and Li, F. 2017: Large-scale analysis reveals that the genome features of simple sequence repeats are generally conserved at the family level in insects: BMC Genomics: 18: 1–10: 10.1186/s12864-017-4234-0.
Du, L., Zhang, C., Liu, Q., Zhang, X., and Yue, B. 2018: Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design: Bioinformatics: 34: 681–683: 10.1093/bioinformatics/btx665.
Duhan, N., Kaur, S., and Kaundal, R. 2023: ranchSATdb: A Genome-Wide Simple Sequence Repeat (SSR) Markers Database of Livestock Species for Mutant Germplasm Characterization and Improving Farm Animal Health: Genes: 14: 1481: 10.3390/genes14071481.
Giamouri, E., Zisis, F., Mitsiopoulou, C., Christodoulou, C., Pappas, A.C., Simitzis, P.E., Kamilaris, C., Galliou, F., Manios, T., Mavrommatis, A., and Tsiplakou, E. 2023: Sustainable Strategies for Greenhouse Gas Emission Reduction in Small Ruminants Farming: Sustainability (Switzerland): 15: 4118: 10.3390/su15054118.
Gochi, L., Kawai, Y., and Fujimoto, A. 2023: Comprehensive analysis of microsatellite polymorphisms in human populations: Human Genetics: 142: 45–57: 10.1007/s00439-022-02484-3.
Hristakieva, P. 2021: Current state of turkey meat production in the world and in Bulgaria.: Bulgarian Journal of Animal Husbandry/Životnov Dni Nauki: 58: 42–49.
Huang, H.B., Song, Y.Q., Hsei, M., Zahorchak, R., Chiu, J., Teuscher, C., and Smith, E.J. 1999: Development and characterization of genetic mapping resources for the turkey (Meleagris gallopavo): Journal of Heredity: 90: 240–242: 10.1093/jhered/90.1.240.
Kheiralipour, K., Rafiee, S., Karimi, M., Nadimi, M., and Paliwal, J. 2024: The environmental impacts of commercial poultry production systems using life cycle assessment: a review: World’s Poultry Science Journal: 80: 33–54: 10.1080/00439339.2023.2250326.
Kumpatla, S.P., and Mukhopadhyay, S. 2005: Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species: Genome: 48: 985–998: 10.1139/g05-060.
Liu, S., Hou, W., Sun, T., Xu, Y., Li, P., Yue, B., Fan, Z., and Li, J. 2017: Genome-wide mining and comparative analysis of microsatellites in three macaque species: Molecular Genetics and Genomics: 292: 537–550: 10.1007/s00438-017-1289-1.
Olubunmi, O.O. 2019: Application of Microsatellite in Fish Biotechnology: Prospects and Drawback-Review: Olagunju Oluwatosin Olubunmi. Application of Microsatellite in Fish Biotechnology: Prospects and Drawback-Review. International Journal of Bioengineering & Biotechnology: 4: 37–43.
Qi, W.H., Jiang, X.M., Du, L.M., Xiao, G.S., Hu, T.Z., Yue, B.S., and Quan, Q.M. 2015: Genome-wide survey and analysis of microsatellite sequences in bovid species: PLoS ONE: 10: e0133667: 10.1371/journal.pone.0133667.
Reed, K.M., Chaves, L.D., and Mendoza, K.M. 2007: An integrated and comparative genetic map of the turkey genome: Cytogenetic and Genome Research: 119: 113–126: 10.1159/000109627.
Safaa, H., Khaled, R., Isaac, S., Mostafa, R., Ragab, M., Elsayed, D.A.A., and Helal, M. 2023: Genome-wide in silico characterization, validation, and cross-species transferability of microsatellite markers in Mallard and Muscovy ducks: Journal of Genetic Engineering and Biotechnology: 21: 105: 10.1186/s43141-023-00556-z.
Thornton, E.K., Emery, K.F., Steadman, D.W., Speller, C., Matheny, R., and Yang, D. 2012: Earliest Mexican Turkeys (Meleagris gallopavo) in the Maya region: Implications for pre-hispanic animal trade and the timing of Turkey domestication: PLoS ONE: 7: e42630: 10.1371/journal.pone.0042630.
Vergara, A.M.C., Landi, V., Bermejo, J.V.D., Martínez, A., Acosta, P.C., Barro, Á.P., Bigi, D., Sponenberg, P., Helal, M., Banabazi, M.H., and Vallejo, M.E.C. 2019: Tracing worldwide Turkey genetic diversity using D-loop sequence mitochondrial DNA analysis: Animals: 9: 10.3390/ani9110897.
Vieira, M.L.C., Santini, L., Diniz, A.L., and Munhoz, C. de F. 2016: Microsatellite markers: What they mean and why they are so useful: Genetics and Molecular Biology: 39: 312–328: 10.1590/1678-4685-GMB-2016-0027.
Wattanadilokchatkun, P., Panthum, T., Jaisamut, K., Ahmad, S.F., Dokkaew, S., Muangmai, N., Duengkae, P., Singchat, W., and Srikulnath, K. 2022: Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity: Fishes: 7: 251: 10.3390/fishes7050251.
Wöhrmann, T., and Weising, K. 2011: In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae: Theoretical and Applied Genetics: 123: 635–647: 10.1007/s00122-011-1613-9.
Zhao, X., Tan, Z., Feng, H., Yang, R., Li, M., Jiang, J., Shen, G., and Yu, R. 2011: Microsatellites in different Potyvirus genomes: Survey and analysis: Gene: 488: 52–56: 10.1016/j.gene.2011.08.016.
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2025

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-CompartirIgual 4.0.
La revista Biotecnia se encuentra bajo la licencia Atribución-NoComercial-CompartirIgual 4.0 Internacional (CC BY-NC-SA 4.0)













_(2).jpg)








