Whole-Genome Characterization and Analysis of Microsatellites in Turkey (Meleagris gallopavo) through in silico Approach
DOI:
https://doi.org/10.18633/biotecnia.v27.2455Keywords:
In silico, Meleagris gallopavo, Microsatellites, Motifs, TurkeysAbstract
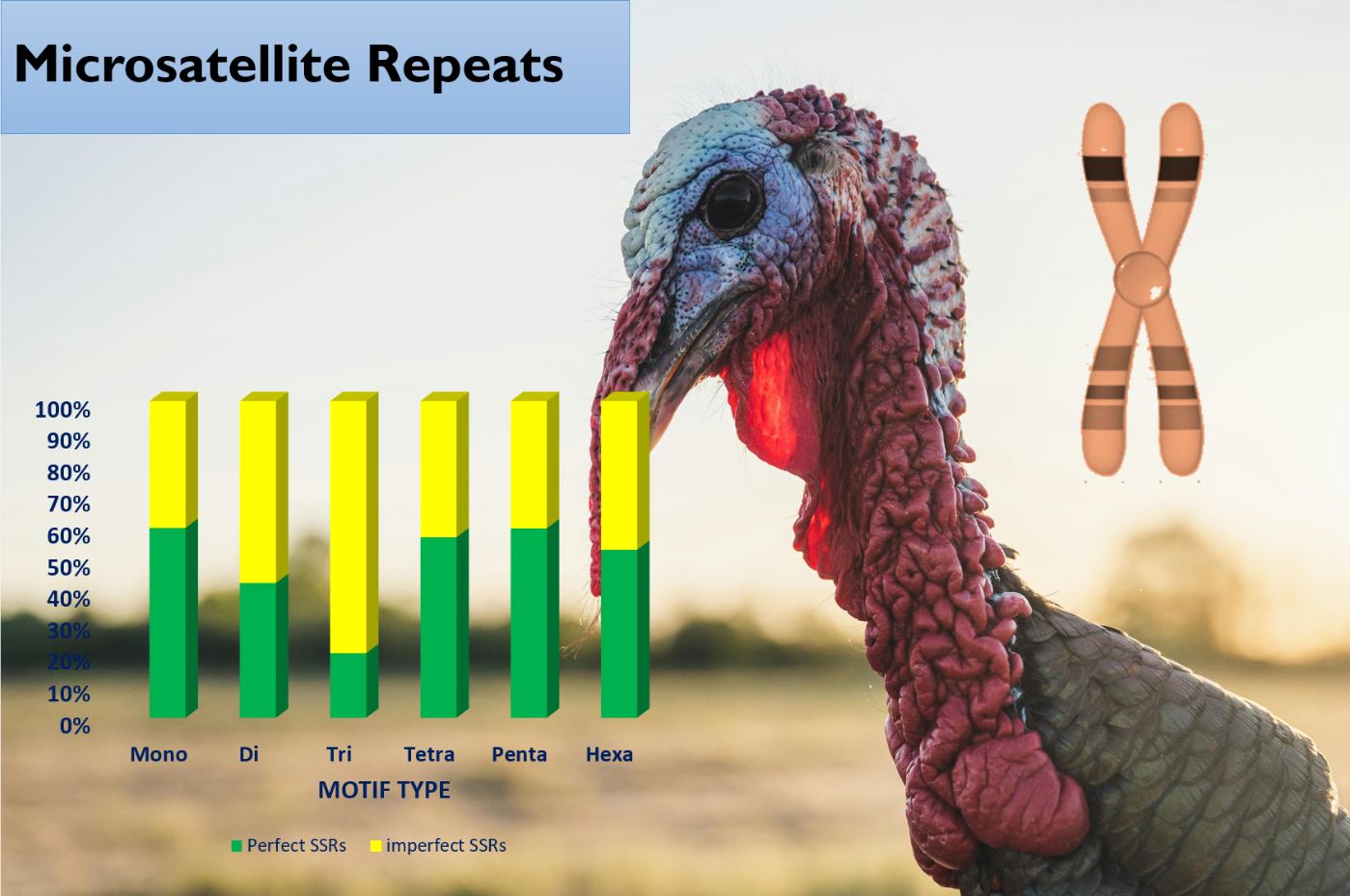
Turkey (Meleagris gallopavo) is the second largest contributor in poultry meat production after chickens. The study of microsatellite organization and distribution is of highly important in genomics and evolutionary students. The in silico mining for microsatellites leverages the power of computational biology to streamline and enhance the discovery of microsatellite markers, and reduce the cost of microsatellite detection. This study aimed at in silico mining microsatellite loci in the genome of domestic turkey. Reference sequences of the different chromosome obtained from NBCI and analyzed using Krait software. Chromosome 4 had the highest number of perfect microsatellites, while chromosome 18 had the lowest. However, chromosome 27 had the highest relative abundance, followed by chromosome. Chromosome 18 again had the lowest relative abundance. For imperfect microsatellites, chromosome 4 had the most imperfect microsatellites and chromosome 18 had the least. A total of 121248 SSR primers were designed. These SSR loci and markers will be instrumental for linkage mapping and significantly improve research on turkey population genetics.
Downloads
References
Abdul-Muneer, P.M. 2014: Application of Microsatellite Markers in Conservation Genetics and Fisheries Management: Recent Advances in Population Structure Analysis and Conservation Strategies: Genetics Research International: 2014: 1–11: 10.1155/2014/691759.
Akemi, A., Pereira, J., Macedo, P., and Alessandra, K. 2012: Microsatellites as Tools for Genetic Diversity Analysis: Genetic Diversity in Microorganisms: 2: 64: 10.5772/35363.
Axelsson, E., Webster, M.T., Smith, N.G.C., Burt, D.W., and Ellegren, H. 2005: Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes: Genome Research: 15: 120–125: 10.1101/gr.3021305.
Barros, C.P., Derks, M.F.L., Mohr, J., Wood, B.J., Crooijmans, R.P.M.A., Megens, H.J., Bink, M.C.A.M., and Groenen, M.A.M. 2023: A new haplotype-resolved turkey genome to enable turkey genetics and genomics research: GigaScience: 12: giad051: 10.1093/gigascience/giad051.
Burt, D.W., Morrice, D.R., Sewalem, A., Smith, J., Paton, I.R., Smith, E.J., Bentley, J., and Hocking, P.M. 2003: Preliminary linkage map of the Turkey (Meleagris gallopavo) based on microsatellite markers: Animal Genetics: 34: 399–409: 10.1046/j.1365-2052.2003.01033.x.
Canales Vergara, A.M., Landi, V., Delgado Bermejo, J.V., Martínez, A.M., Cervantes Acosta, P., Pons Barros, A., Bigi, D., Sponenberg, P., Helal, M., Banabazi, M.H., and Camacho Vallejo, M.E. 2020: Design and development of a multiplex microsatellite panel for the genetic characterisation and diversity assessment of domestic turkey (Meleagris gallopavo gallopavo): Italian Journal of Animal Science: 19: 392–398: 10.1080/1828051X.2020.1745695.
Clauss, M., Frei, S., Hatt, J.M., and Kreuzer, M. 2020: Methane emissions of geese (Anser anser) and turkeys (Meleagris gallopavo) fed pelleted lucerne: Comparative Biochemistry and Physiology -Part A : Molecular and Integrative Physiology: 242: 110651: 10.1016/j.cbpa.2020.110651.
Dalloul, R.A., Long, J.A., Zimin, A. V., Aslam, L., Beal, K., Blomberg, L.A., Bouffard, P., Burt, D.W., Crasta, O., Crooijmans, R.P.M.A., Cooper, K., Coulombe, R.A., De, S., Delany, M.E., Dodgson, J.B., Dong, J.J., Evans, C., Frederickson, K.M., Flicek, P., Florea, L., Folkerts, O., Groenen, M.A.M., Harkins, T.T., Herrero, J., Hoffmann, S., Megens, H.J., Jiang, A., de Jong, P., Kaiser, P., Kim, H., Kim, K.W., Kim, S., Langenberger, D., Lee, M.K., Lee, T., Mane, S., Marcais, G., Marz, M., McElroy, A.P., Modise, T., Nefedov, M., Notredame, C., Paton, I.R., Payne, W.S., Pertea, G., Prickett, D., Puiu, D., Qioa, D., Raineri, E., Ruffier, M., Salzberg, S.L., Schatz, M.C., Scheuring, C., Schmidt, C.J., Schroeder, S., Searle, S.M.J., Smith, E.J., Smith, J., Sonstegard, T.S., Stadler, P.F., Tafer, H., Tu, Z., van Tassell, C.P., Vilella, A.J., Williams, K.P., Yorke, J.A., Zhang, L., Zhang, H. Bin, Zhang, X., Zhang, Y., and Reed, K.M. 2010: Multi-platform next-generation sequencing of the domestic Turkey (Meleagris gallopavo): Genome assembly and analysis: PLoS Biology: 8: e1000475: 10.1371/journal.pbio.1000475.
Ding, S., Wang, S., He, K., Jiang, M., and Li, F. 2017: Large-scale analysis reveals that the genome features of simple sequence repeats are generally conserved at the family level in insects: BMC Genomics: 18: 1–10: 10.1186/s12864-017-4234-0.
Du, L., Zhang, C., Liu, Q., Zhang, X., and Yue, B. 2018: Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design: Bioinformatics: 34: 681–683: 10.1093/bioinformatics/btx665.
Duhan, N., Kaur, S., and Kaundal, R. 2023: ranchSATdb: A Genome-Wide Simple Sequence Repeat (SSR) Markers Database of Livestock Species for Mutant Germplasm Characterization and Improving Farm Animal Health: Genes: 14: 1481: 10.3390/genes14071481.
Giamouri, E., Zisis, F., Mitsiopoulou, C., Christodoulou, C., Pappas, A.C., Simitzis, P.E., Kamilaris, C., Galliou, F., Manios, T., Mavrommatis, A., and Tsiplakou, E. 2023: Sustainable Strategies for Greenhouse Gas Emission Reduction in Small Ruminants Farming: Sustainability (Switzerland): 15: 4118: 10.3390/su15054118.
Gochi, L., Kawai, Y., and Fujimoto, A. 2023: Comprehensive analysis of microsatellite polymorphisms in human populations: Human Genetics: 142: 45–57: 10.1007/s00439-022-02484-3.
Hristakieva, P. 2021: Current state of turkey meat production in the world and in Bulgaria.: Bulgarian Journal of Animal Husbandry/Životnov Dni Nauki: 58: 42–49.
Huang, H.B., Song, Y.Q., Hsei, M., Zahorchak, R., Chiu, J., Teuscher, C., and Smith, E.J. 1999: Development and characterization of genetic mapping resources for the turkey (Meleagris gallopavo): Journal of Heredity: 90: 240–242: 10.1093/jhered/90.1.240.
Kheiralipour, K., Rafiee, S., Karimi, M., Nadimi, M., and Paliwal, J. 2024: The environmental impacts of commercial poultry production systems using life cycle assessment: a review: World’s Poultry Science Journal: 80: 33–54: 10.1080/00439339.2023.2250326.
Kumpatla, S.P., and Mukhopadhyay, S. 2005: Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species: Genome: 48: 985–998: 10.1139/g05-060.
Liu, S., Hou, W., Sun, T., Xu, Y., Li, P., Yue, B., Fan, Z., and Li, J. 2017: Genome-wide mining and comparative analysis of microsatellites in three macaque species: Molecular Genetics and Genomics: 292: 537–550: 10.1007/s00438-017-1289-1.
Olubunmi, O.O. 2019: Application of Microsatellite in Fish Biotechnology: Prospects and Drawback-Review: Olagunju Oluwatosin Olubunmi. Application of Microsatellite in Fish Biotechnology: Prospects and Drawback-Review. International Journal of Bioengineering & Biotechnology: 4: 37–43.
Qi, W.H., Jiang, X.M., Du, L.M., Xiao, G.S., Hu, T.Z., Yue, B.S., and Quan, Q.M. 2015: Genome-wide survey and analysis of microsatellite sequences in bovid species: PLoS ONE: 10: e0133667: 10.1371/journal.pone.0133667.
Reed, K.M., Chaves, L.D., and Mendoza, K.M. 2007: An integrated and comparative genetic map of the turkey genome: Cytogenetic and Genome Research: 119: 113–126: 10.1159/000109627.
Safaa, H., Khaled, R., Isaac, S., Mostafa, R., Ragab, M., Elsayed, D.A.A., and Helal, M. 2023: Genome-wide in silico characterization, validation, and cross-species transferability of microsatellite markers in Mallard and Muscovy ducks: Journal of Genetic Engineering and Biotechnology: 21: 105: 10.1186/s43141-023-00556-z.
Thornton, E.K., Emery, K.F., Steadman, D.W., Speller, C., Matheny, R., and Yang, D. 2012: Earliest Mexican Turkeys (Meleagris gallopavo) in the Maya region: Implications for pre-hispanic animal trade and the timing of Turkey domestication: PLoS ONE: 7: e42630: 10.1371/journal.pone.0042630.
Vergara, A.M.C., Landi, V., Bermejo, J.V.D., Martínez, A., Acosta, P.C., Barro, Á.P., Bigi, D., Sponenberg, P., Helal, M., Banabazi, M.H., and Vallejo, M.E.C. 2019: Tracing worldwide Turkey genetic diversity using D-loop sequence mitochondrial DNA analysis: Animals: 9: 10.3390/ani9110897.
Vieira, M.L.C., Santini, L., Diniz, A.L., and Munhoz, C. de F. 2016: Microsatellite markers: What they mean and why they are so useful: Genetics and Molecular Biology: 39: 312–328: 10.1590/1678-4685-GMB-2016-0027.
Wattanadilokchatkun, P., Panthum, T., Jaisamut, K., Ahmad, S.F., Dokkaew, S., Muangmai, N., Duengkae, P., Singchat, W., and Srikulnath, K. 2022: Characterization of Microsatellite Distribution in Siamese Fighting Fish Genome to Promote Conservation and Genetic Diversity: Fishes: 7: 251: 10.3390/fishes7050251.
Wöhrmann, T., and Weising, K. 2011: In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae: Theoretical and Applied Genetics: 123: 635–647: 10.1007/s00122-011-1613-9.
Zhao, X., Tan, Z., Feng, H., Yang, R., Li, M., Jiang, J., Shen, G., and Yu, R. 2011: Microsatellites in different Potyvirus genomes: Survey and analysis: Gene: 488: 52–56: 10.1016/j.gene.2011.08.016.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
The journal Biotecnia is licensed under the Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0) license.














_(2).jpg)







