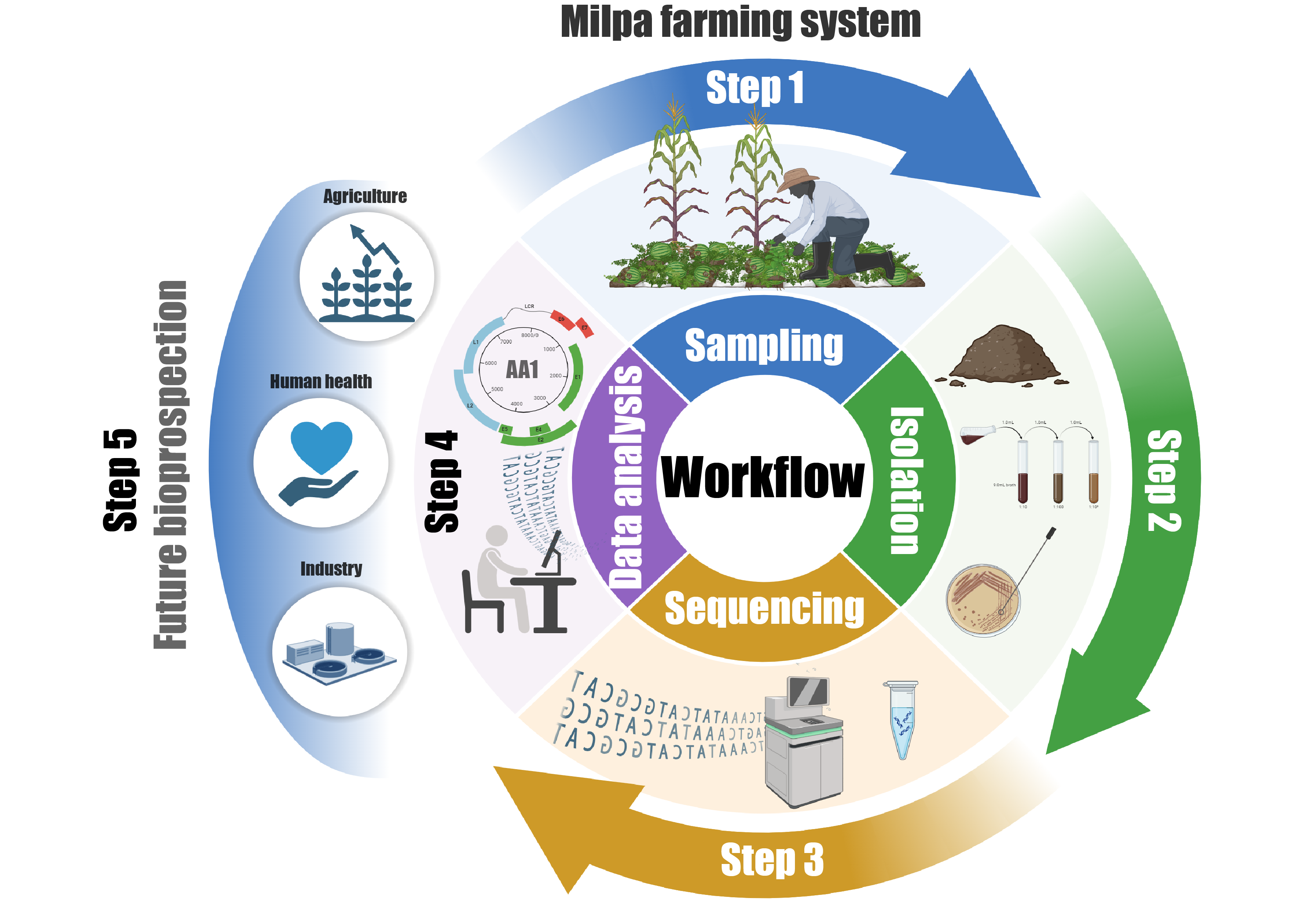
Genome Analysis of Bacillus paralicheniformis AA1 isolated from a conventional milpa farming system in the northwestern region of Sonora, Mexico
DOI:
https://doi.org/10.18633/biotecnia.v27.2552Keywords:
Bacillus paralicheniformis, genoma completo, agricultura sostenibleAbstract
This study provides an in-depth genome analysis of Bacillus
paralicheniformis AA1, a bacterial strain isolated from a
traditional milpa farming system in Sonora, Mexico. The
genomic analysis revealed a high level of completeness,
demonstrated by the presence of a diverse and functionally
significant repertoire of genes associated with fundamental
biological processes, including nutrient assimilation, stress
response, and cellular regulation. Notably, the genome also
contains genes responsible for the biosynthesis of secondary metabolites, highlighting its potential for biotechnological applications. Taxonomic classification was rigorously conducted using integrated genome-wide approaches, which definitively confirmed the identification of iso late AA1 as belonging to the species Bacillus paralicheniformis. Comparative genomic analysis further established a high degree of genetic similarity between AA1 and other B. paralicheniformis strains with well-characterized biotechnological capabilities. This similarity strongly suggests that AA1 harbors genetic elements responsible for the synthesis of antimicrobial compounds, enzymes with industrial relevance, and metabolites that promote plant growth. The findings underscore the potential of Bacillus paralicheniformis AA1 as a valuable resource for biotechnology and sustainable agriculture. By enhancing our understanding of microbial diversity within traditional agroecosystems, this study contributes to the broader knowledge base required for the development of innovative agricultural practices. Future research should focus on functional validation of key genes to fully unlock AA1’s potential as a bioresource for antimicrobial production, enzyme synthesis, and crop enhancement, paving the way for its application in environmentally sustainable farming systems.
Downloads
References
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N. 2018. A global atlas of the dominant bacteria found in soil. Science (1979) 359:320–325.
Steinke K, Mohite OS, Weber T, Kovács ÁT. 2021. Phylogenetic Distribution of Secondary Metabolites in the Bacillus subtilis Species Complex. mSystems 6.
Su Y, Liu C, Fang H, Zhang D. 2020. Bacillus subtilis: a universal cell factory for industry, agriculture, biomaterials and medicine. Microbial Cell Factories 2020 19:1 19:1–12.
Iqbal S, Qasim M, Rahman H, Khan N, Paracha RZ, Bhatti MF, Javed A, Janjua HA. 2023. Genome mining, antimicrobial and plant growth-promoting potentials of halotolerant Bacillus paralicheniformis ES-1 isolated from salt mine. Molecular Genetics and Genomics 298:79–93.
Ashajyothi M, Mahadevakumar S, Venkatesh YN, Sarma PVSRN, Danteswari C, Balamurugan A, Prakash G, Khandelwal V, Tarasatyavathi C, Podile AR, Kirankumar MS, Chandranayaka S. 2024. Comprehensive genomic analysis of Bacillus subtilis and Bacillus paralicheniformis associated with the pearl millet panicle reveals their antimicrobial potential against important plant pathogens. BMC Plant Biol 24:1–23.
Swietczak J, Kalwasińska A, Felföldi T, Swiontek Brzezinska M. 2023. Bacillus paralicheniformis 2R5 and its impact on canola growth and N-cycle genes in the rhizosphere. FEMS Microbiol Ecol 99.
Chavarria-Quicaño E, Contreras-Jácquez V, Carrillo-Fasio A, De la Torre-González F, Asaff-Torres A. 2023. Native Bacillus paralicheniformis isolate as a potential agent for phytopathogenic nematodes control. Front Microbiol 14.
Jinal HN, Sakthivel K, Amaresan N. 2020. Characterisation of antagonistic Bacillus paralicheniformis (strain EAL) by LC–MS, antimicrobial peptide genes, and ISR determinants. Antonie van Leeuwenhoek, International Journal of General and Molecular Microbiology 113:1167–1177.
Díaz-Manzano FE, Amora DX, Martínez-Gómez Á, Moelbak L, Escobar C. 2023. Biocontrol of Meloidogyne spp. in Solanum lycopersicum using a dual combination of Bacillus strains. Front Plant Sci 13:1077062.
Hemangini BD, Kiran CS, Vrinda TS, Bhatt Hemangini CD. 2018. Antimicrobial activity of Bacillus paralicheniformis SUBG0010 against plant pathogenic bacteria of mango. ~ 449 ~ Journal of Pharmacognosy and Phytochemistry 7:449–455.
Pawaskar M. 2023. In-vivo biocontrol of Fusarium wilt in chilli plants using hypersaline Bacillus paralicheniformis strain MPSK23 https://doi.org/10.21203/RS.3.RS-2927693/V1.
Fonteyne S, Castillo Caamal JB, Lopez-Ridaura S, Van Loon J, Espidio Balbuena J, Osorio Alcalá L, Martínez Hernández F, Odjo S, Verhulst N. 2023. Review of agronomic research on the milpa, the traditional polyculture system of Mesoamerica. Frontiers in Agronomy 5:1115490.
Compant S, Samad A, Faist H, Sessitsch A. 2019. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J Adv Res 19:29–37.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin A V., Sirotkin A V., Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology 19:455.
Tatusova T, Dicuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624.
Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, Trigodet F, Watson AR, Esen ÖC, Moore RM, Clayssen Q, Lee MD, Kivenson V, Graham ED, Merrill BD, Karkman A, Blankenberg D, Eppley JM, Sjödin A, Scott JJ, Vázquez-Campos X, McKay LJ, McDaniel EA, Stevens SLR, Anderson RE, Fuessel J, Fernandez-Guerra A, Maignien L, Delmont TO, Willis AD. 2020. Community-led, integrated, reproducible multi-omics with anvi’o. Nature Microbiology 2020 6:1 6:3–6.
Lee MD. 2019. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35:4162–4164.
Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen CY, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res 51:W484–W492.
Klimke W, O’Donovan C, White O, Brister JR, Clark K, Fedorov B, Mizrachi I, Pruitt KD, Tatusova T. 2011. Solving the Problem: Genome Annotation Standards before the Data Deluge. Stand Genomic Sci 5:168.
Berais-Rubio A, Morel-Revetria M, Filippi CV, Reyno R, Monza J. 2023. Ensifer meliloti elite strain U143 used as alfalfa inoculant in Uruguay: Characterization and draft genome sequence. The Microbe 1:100008.
Berriel V, Morel MA, Filippi C V., Monza J. 2021. Draft genome sequence of Bradyrhizobium sp. strain Oc8 isolated from Crotalaria ochroleuca nodule. Curr Res Microb Sci 2:100074.
Kim M, Chun J. 2014. 16S rRNA Gene-Based Identification of Bacteria and Archaea using the EzTaxon Server. Methods in Microbiology 41:61–74.
Bars-Cortina D, Moratalla-Navarro F, García-Serrano A, Mach N, Riobó-Mayo L, Vea-Barbany J, Rius-Sansalvador B, Murcia S, Obón-Santacana M, Moreno V. 2023. Improving Species Level-taxonomic Assignment from 16S rRNA Sequencing Technologies. Curr Protoc 3:e930.
Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil PA, Hugenholtz P. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794.
Arahal DR. 2014. Whole-Genome Analyses: Average Nucleotide Identity. Methods in Microbiology 41:103–122.
Asif M, Li-Qun Z, Zeng Q, Atiq M, Ahmad K, Tariq A, Al-Ansari N, Blom J, Fenske L, Alodaini HA, Hatamleh AA. 2023. Comprehensive genomic analysis of Bacillus paralicheniformis strain BP9, pan-genomic and genetic basis of biocontrol mechanism. Comput Struct Biotechnol J 21:4647–4662.
Annapurna K, Govindasamy V, Sharma M, Ghosh A, Chikara SK. 2018. Whole genome shotgun sequence of Bacillus paralicheniformis strain KMS 80, a rhizobacterial endophyte isolated from rice (Oryza sativa L.). 3 Biotech 8:1–4.
Valenzuela-Ruiz V, Robles-Montoya RI, Parra-Cota FI, Santoyo G, del Carmen Orozco-Mosqueda M, Rodríguez-Ramírez R, de los Santos-Villalobos S. 2019. Draft genome sequence of Bacillus paralicheniformis TRQ65, a biological control agent and plant growth-promoting bacterium isolated from wheat (Triticum turgidum subsp. durum) rhizosphere in the Yaqui Valley, Mexico. 3 Biotech 9:1–7.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
The journal Biotecnia is licensed under the Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0) license.














_(2).jpg)







