Isolated and identification of yeast of "Palo podrido" degradation with cellulolytic, xylanolytic, and laccase enzymatic activity
Isolation and identification of yeasts
Keywords:
Cellulase, Laccase, Lignocellulolytic yeasy, Palo podrido, XylanaseAbstract
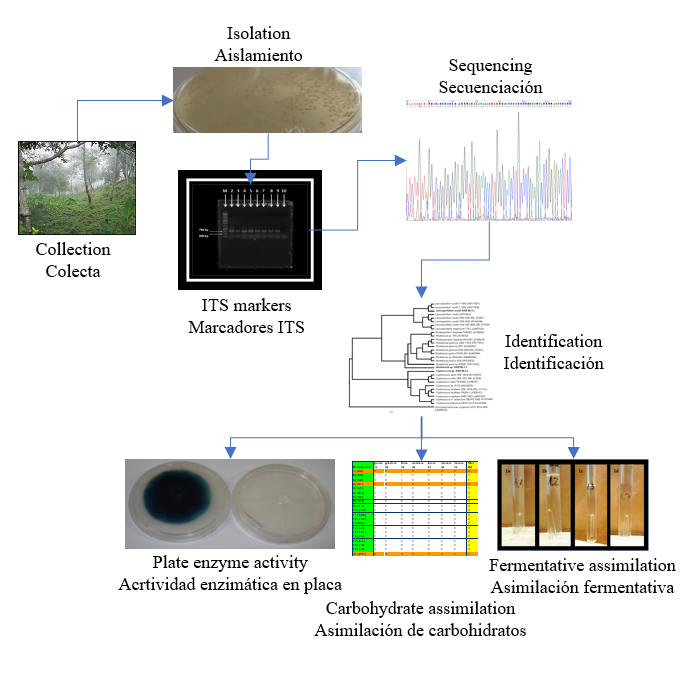
Ten yeasts were isolated from samples obtained from the degradation of wood called "Palo podrido". The yeasts were isolated and identified from the sequencing of the ITS fragment. The phylogenetic analysis identified three strains of basidiomycetes of the genera Rhodotorula, Cryptococcus, and Leucosporidium and seven strains of ascomycetes of the genera Candida, Sugiyamaella, and Spencermartinsiella. Carbohydrate assimilation studies were carried out, and none of the isolated strains could assimilate starch; all strains assimilated glucose and xylose. Cryptococcus sp., Rhodotorula sp., and Leucosporidum sp. showed fermentative metabolism. Rhodotorula sp., Cryptococcus sp., Leucospororidium sp., Candida sp., and Sugiyamaella sp. showed both cellulolytic and xylanolytic activity on a plate. The hydrolytic enzymatic activity was influenced by the type and nitrogen concentration. The strains that showed cellulolytic and xylanolytic activity were dependent on the type and nitrogen concentration in the culture medium; only Rhodotorula sp. presented hydrolytic activity at three concentrations of total nitrogen (1, 2, and 3 %) and with only one type of nitrogen source. For its part, Cryptococcus sp. was the only strain that showed laccase activity in a medium supplemented with glycerol as a carbon source.
Downloads
References
Arana, A., Téllez, A., González, T., González. A. 2002. Aspectos generales de la biodegradación de la madera: aplicaciones industriales de las lacasas. BioTecnología. 7(3):40-55.
Baffi, M.A., Bezerra, C.S., Arevalo-Villena, M., Briones-Pérez, A.I., Gomes, E., Da Silva, R. 2011. Isolation and molecular identification of wine yeasts from a Brazilian vineyard. Annals Microbiology. 61: 75–78.
Bastawde, K.B., Puntambekar, U.S., Gokhale, D.V. 1994. Optimization of cellulase-free xylanase production by a novel yeast strain. Journal of Industrial Microbiology and Biotechnology. 13(4): 220-224.
Betini, J.H.A., Michelin, M., Peixoto-Nogueira, S.C., Jorge, J.A., Terenzi, H.F., Polizeli, M.L.T.M. 2009. Xylanases from Aspergillus niger, Aspergillus niveus and Aspergillus ochraceus produced under solid-state fermentation and their application in cellulose pulp bleaching. Bioprocess and Biosystems Engineering. 32: 819-24.
Blanchette, R.A., Shaw, C.G. 1978. Association among bacteria, yeast, and basidiomycetes durin wood decay. Phytopathology. 68: 631-637.
Brandão, L.R., Libkind, D., Vaz, A.B.M., Espírito Santo, L.C., Moliné, M., de García, V., van Broock, M., Rosa, C.A. 2011. Yeasts from an oligotrophic lake in Patagonia (Argentina): diversity, distribution and synthesis of photoprotective compounds and extracellular enzymes. FEMS Microbiology Ecology. 76: 1-13.
Brooks, A.A. 2008. Ethanol production potential of local yeast strains isolated from ripe banana peels. African Journal of Biotechnology. 7: 20.
Bura, R., Vajzovic, A., Doty, S.L. 2012. Novel endophytic yeast Rhodotorula mucilaginosa strain PTD3 I: production of xylitol and ethanol. Journal of Industrial Microbiology and Biotechnology. 39(7): 1003-1011.
Cadete, R.M., Melo, M.A., Lopes, M.R., Pereira, G.M.D., Zilli, J.E., Vital, M.J.S., Gomes, F.C.O., Lachance, M.A., Rosa, C.A. 2012. Candida amazonensis sp. nov., an ascomycetous yeast isolated from rotting wood in the Amazonian forest. International Journal of Systematic and Evolutionary Microbiology. 62: 1438-1440.
Carrasco, M., Rozas, J.M., Barahona, S., Alcaíno, J., Cifuentes, V., Baeza, M. 2012. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiology. 12(1): 1-9.
Chang, Y.H., Chang, K.S., Lee, C.F., Hsu, C.L., Huang, C.W., Jang, H.D. 2015. Microbial lipid production by oleaginous yeast Cryptococcus sp. in the batch cultures using corncob hydrolysate as carbon source. Biomass and Bioenergy. 72: 95-103.
Chen, S.H., Williamson, P.R. 2011. Lessons from Cryptococcal Laccase: From Environmental Saprophyte to Pathogen. Current Fungal Infection Reports. 5: 233-244.
Cruz-Ramírez, M.G., Rivera-Ríos, J.M., Téllez-Jurado, A., Maqueda-Gálvez, A.P., Mercado-Flores, Y., Arana-Cuenca, A. 2012. Screening for thermotolerant ligninolytic fungi with laccase, lipase, and protease activity isolated in Mexico. Journal of Environmental Management. 95: S256-S259.
De Mot, R., Van Dijck, K., Donkers, A., Verachtert, H. 1985. Potentialities and limitations of direct alcoholic fermentation of starchy material with amylolytic yeasts. Applied Microbiology and Biotechnology. 22(3): 222-226.
Diamantopoulou, P., Stoforos, N.G., Xenopoulos, E., Sarris, D., Psarianos, D., Philippoussis, A., Papanikolaou, S. 2020. Lipid production by Cryptococcus curvatus growing on commercial xylose and subsequent valorization of fermentation waste-waters for the production of edible and medicinal mushrooms. Biochemical Engineering Journal. 162: 107706.
Duarte, A.W.F., Dayo-Owoyemi, I., Nobre, F.S., Pagnocca, F.C., Chaud, L.C.S., Pessoa, A., Felipe, M.G.A., Sette, L.D. 2013. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles. 17(6): 1023-1035.
Elisashvili, V., Kachlishvili, E., Penninckx, M. 2008. Effect of growth substrate, method of fermentation, and nitrogen source on lignocellulose-degrading enzymes production by white-rot basidiomycetes. Journal of Industrial Microbiology and Biotechnology. 35(11): 1531-1538.
Edwards, I.P., Zak, D.R., Kellner, H., Eisenlord, S.D., Pregitzer, K.S. 2011. Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PloS One. 6(6): e20421.
Flores, C.L., Rodríguez, C., Petit, T., Gacendo, C. 2000. Carbohydrate and energy-yielding metabolism in non-conventional yeast. FEMS Microbiology Reviews. 24: 507-509.
Gientka, I., Bzducha-Wróbel, A., Stasiak-Różańska, L., Bednarska, A.A., Błażejak, S. 2016. The exopolysaccharides biosynthesis by Candida yeast depends on carbon sources. Electronic Journal of Biotechnology. 22: 31-37.
Gomes, J., Gomes, I., Esterbauer, H., Kreiner, W., Steiner, W. 1989 Production of cellulases by a wild strain of Gliocladium virens: optimization of the fermentation medium and partial characterization of the enzymes. Applied Microbiology and Biotechnology. 31: 601-608.
Gosalawit, C., Imsoonthornruksa, S., Gilroyed, B.H., Mcnea, L., Boontawan, A., Ketudat-Cairns, M. 2021. The potential of the oleaginous yeast Rhodotorula paludigena CM33 to produce biolipids. Journal of Biotechnology. 329: 56-64.
González, A.E., Martínez, A.T., Almendros, G., Grinbergs, J. 1989. A study of yeasts during the delignification and fungal transformation of wood into cattle feed in Chilean rain forest. Antonie van Leeuwenhoek. 55: 221-236.
Grotjohan, N., Kowallik, W., Huang, Y. 2000. Investigations into enzymes of nitrogen metabolism of the ectomycorrhizal basidiomycete, Suillus bovinus. Zeitschrift für Naturforschung C. 55(3-4): 203-212.
Guo, X., Zhu, H., Bai, F.Y. 2012. Candida cellulosicola sp. nov., a xylose-utilizing anamorphic yeast from rotten wood. International Journal of Systematic and Evolutionary Microbiology. 62: 242-245.
Haapala, R., Parkkinen, E., Linko, S. 1996. Production of endo-1,4-β-glucanase and xylanase with nylon- web immobilized and free Trichoderma reesei. Enzyme and Microbial Technology. 18: 495-501.
Hamidi, M., Gholipour, A.R., Delattre, C., Sesdighi, F., Seveiri, R.M., Pasdaran, A., Kheirandish, S., Pierre, G., Kozani, P.S., Kozani, P.S., Karimitabar, F. 2020. Production, characterization and biological activities of exopolysaccharides from a new cold-adapted yeast: Rhodotorula mucilaginosa sp. GUMS16. International Journal of Biological Macromolecules. 151: 268-277.
Jiménez, M., González, A.E., Martínez, M.J., Martínez, A.T., Dale, B.E. 1991. Screening of yeasts isolated from decayed wood for lignocellulose-degrading enzymatic activities. Mycological Research. 95: 1299-1302.
Johnson, E.A. 2013. Biotechnology of non-Saccharomyces yeasts-the basidiomycetes. Applied Microbiology and Biotechnology. 97: 7563-7577.
Kurtzman, C.P. 2011. Discussion of teleomorphic and anamorphic ascomycetous yeasts and yeast-like taxa. Ch 13. In The yeasts (pp. 293-307). Elsevier.
Lara, C.A., Santos, R.O., Cadete, R.M., Ferreira, C., Marques, S., Gírio, F., Oliveira, E.S., Rosa, C.A., Fonseca, C. 2014. Identification and characterisation of xylanolytic yeasts isolated from decaying wood and sugarcane bagasse in Brazil. Antonie van Leeuwenhoek. 105: 1107 -1119.
Lazera, M.S., Pires, F.D., Camillo-Coura, L., Nishikawa, M.M., Bezerra, C.C., Trilles, L., Wanke, B. 1996. Natural habitat of Cryptococcus neoformans var. neoformans in decaying wood forming hollows in living trees. Journal of Medical and Veterinary Mycology. 34: 127-131.
Lee, K.M., Kalyani, D., Tiwari, M.K., Kim, T.S., Dhiman, S.S., Lee, J.K., Kim, I.W. 2012. Enhanced enzymatic hydrolysis of rice straw by removal of phenolic compounds using a novel laccase from yeast Yarrowia lipolytica. Bioresource Technology. 123: 636-645.
Liang, Y., Feng, Z., Yesuf, J., Blackburn, J.W. 2010. Optimization of growth medium and enzyme assay conditions for crude cellulases produced by a novel thermophilic and cellulolytic bacterium, Anoxybacillus sp. 527. Applied Biochemistry and Biotechnology. 160:1 841-52.
Maaheimo, H., Fiaux, J., Cakar, Z.P., Bailey, J.E., Sauer, U., Szyperski, T. 2002. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional (13)C labeling of common amino acids. European Journal of Biochemistry 268: 2464-2479.
Martins, G.M., Bocchini-Martins, D.A., Bezzerra-Bussoli, C., Pagnocca, F.C., Boscolo, M., Monteiro, D.A., Gomes, E. 2018 The isolation of pentose-assimilating yeasts and their xylose fermentation potential. Brazilian Journal of Microbiology. 49(1): 162-168.
Martius, C. 1992. Density, humidity, and nitrogen content of dominant wood species of floodplain forests (várzea) in Amazonia. Holz als Roh und Werkstoff. 50(7-8): 300-303.
Mazza, M., Refojo, N., Bosco-Borgeat, M.E., Taverna, C.G., Trovero, A.C., Rogé, A., Davel, G. 2013. Cryptococcus gattii in urban trees from cities in North-eastern Argentina. Mycoses 56: 646-650.
Morais, C.G., Cadete, R.M., Uetanabaro, A.P.T., Rosa, L.H., Lachance, M.A., Rosa, C.A. 2013. D-xylose-fermenting and xylanase-producing yeast species from rotting wood of two Atlantic Rainforest habitats in Brazil. Fungal Genetics and Biology. 60: 19-28.
Morais, C.G., Sena, L.M., Lopes, M.R., Santos, A.R.O., Barros, K.O., Alves, C.R., Uetanabaro, A.P.T., Lachance, M.A., Rosa, C.A. 2020. Production of ethanol and xylanolytic enzymes by yeasts inhabiting rotting wood isolated in sugarcane bagasse hydrolysate. Fungal Biology. 124(7): 639-647.
Ortiz, R., Párraga, M., Navarrete, J., Carrasco, I., de la Vega, E., Ortiz, M., Herrera, P., Jurgens, J.A., Held, B.W., Blanchette, R.A. 2014. Investigations of Biodeterioration by Fungi in Historic Wooden Churches of Chiloé, Chile. Microbial Ecology. 67: 568-575.
Péter, G., Dlauchy, D., Tornai-Lehoczki, J., Suzuki, M., Kurtzman, C. 2011. Spencermartinsiella europaea gen. nov., sp. nov., a new member of the family Trichomonas caceae. International Journal of Systematic and Evolutionary Microbiology. 61: 993-1000.
Phitsuwan, P., Laohakunjit, N., Kerdchoechuen, O., Kyu, K.L., Ratanakhanokchai, K. 2013. Present and potential applications of cellulases in agriculture, biotechnology, and bioenergy. Folia Microbiology. 58: 163-176.
Qadri, S.H., Nichols, C.W. 1978. Tube carbohydrate assimilation method for the rapid identification of clinically significant yeasts. Medical Microbiology and Immunology. 165(1): 19-27.
Raeder, U., Broda, P. 1985. Rapid preparation of DNA from filamentous fungi. Letters in Applied Microbiology. 1: 17– 20.
Ramírez, C., González, A. 1985. Rhodotorula nothofagi sp. nov., isolated from decayed wood in the evergreen rainy Valdivian Forest of southern Chile. Mycopathologia. 91(3): 171-173.
Rani, V., Dash, S., Nain, L., Arora, A. 2015. Expression of novel glucose tolerant β-glucosidase on cell surface by Rhodotorula glutinis isolate. Biocatalysis and Agricultural Biotechnology. 4(3): 380-387.
Ríos, S., Eyzaguirre, J. 1992. Conditions for selective degradation of lignin by the fungus Ganoderma australis. Applied Microbiology and Biotechnology. 37: 667-669.
Sarawan, S., Mahakhan, P., Jindamorakot, S., Vichitphan, K., Vichitphan, S., Sawaengkaew, J. 2013. Candida konsanensis sp. nov., a new yeast species isolated from Jasminum adenophyllum in Thailand with potentially carboxymethyl cellulase-producing capability. World Journal of Microbiology and Biotechnology. 29: 1481-1486.
Shahriarinour, M., Wahab, M.N.A., Mohamad, R., Mustafa, S., Ariff, A.B. 2011. Effect of medium composition and cultural condition on cellulase production by Aspergillus terreus. African Journal of Biotechnology. 10: 7459-7467.
Shariq, M., Sohail, M. 2020. Production of cellulase and xylanase from Candida tropicalis (MK-118) on purified and crude substrates. Pakistan Journal of Botany. 52(1): 323-328.
Sidrim, J.J.C., Rocha, M.F.G., Leite, J.J.G., Maranhão, F.C.D.A., Lima, R.A.C., Castelo-Branco, D.D.S.C.M., Bandeira, T.J.P.G., Cordeiro, R.A., Brilhante, R.S.N. 2013. Trichophyton tonsurans strains from Brazil: phenotypic heterogeneity, genetic homology, and detection of virulence genes. Canadian Journal of Microbiology. 59(11): 754-760.
Singhal, A., Choudhary, G., Thakur, I.S. 2012. Characterization of laccase activity produced by Cryptococcus albidus. Preparative Biochemistry and Biotechnology. 42: (2)113-124.
Takemoto, S., Hwang, W.J., Takeuchi, M., Itoh, T., Imamura, Y. 2008. Anatomical characterization of decayed wood in standing light red meranti and identification of the fungi isolated from the decayed area. Journal of Wood Science. 54: 233-241.
Turkiewicz, M., Pazgier, M., Donachie, S.P., Kalinowska, H. 2005. Invertase and a-glucosidase production by the endemic Antarctic marine yeast Leucosporidium antarcticum. Polish Polar Research. 125-136.
White, T.J., Bruns, T., Lee, S., Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A guide to methods and applications (eds. M.A. Innis, D.H. Gelfand, J.J. Sninsky and T.J. White). Academic Press. USA. 315-332.
Wu, R., Chen, D., Cao, S., Lu, Z., Huang, J., Lu, Q., Chen, Y., Che, X., Guan, N., Wei, Y., Huang, R. 2020. Enhanced ethanol production from sugarcane molasses by industrially engineered Saccharomyces cerevisiae via replacement of the PHO4 gene. RSC Advances. 10(4): 2267-2276.
Yu, X., Zheng, Y., Xiong, X., Chen, S. 2014. Co-utilization of glucose, xylose and cellobiose by the oleaginous yeast Cryptococcus curvatus. Biomass and Bioenergy. 71: 340-349.
Zadrazil, F., Grinbergs, J., González, A.E. 1982. Palo podrido-Decomposed wood which was used as Feed. European Journal of Applied Microbiology and Biotechnology. 15: 167-171.
Zhu, X., Williamson, P.R. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Research. 5(1): 1-10.
Additional Files
Published
How to Cite
Issue
Section
License
Copyright (c) 2023

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
The journal Biotecnia is licensed under the Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0) license.






_(1)_(1).png)






_(2).jpg)




